 |
| berkley.edu |
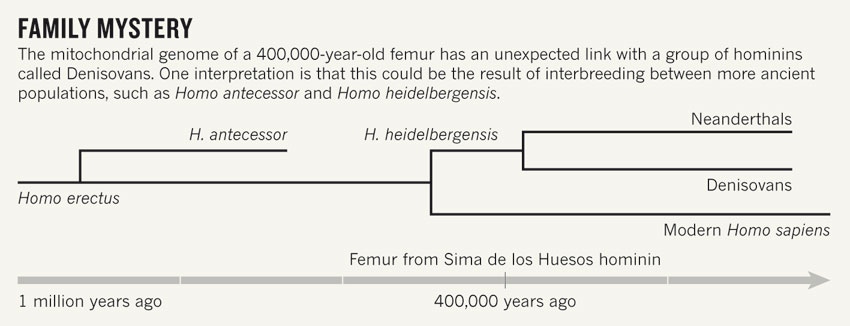
Neanderthals and humans likely diverged from their last common ancestor, which has eluded paleoanthropologists (1), about 1 mya; however, molecular clock data suggests a more recent LCA (2). Neanderthals disappeared around 30,000 years ago, but co-existed with 'humans' for a period of time, and likely interbred somewhere around 47,000-65,000 years ago (3). (i use human loosely as the debate still rages amongst Anthropologists as to whether Neanderthals should be considered 'human'). Because of this interbreeding, anywhere from 1-4% of the genomes from Non-Africans is derived from Neanderthals. Recognizing that some of us are walking around with Neanderthal alleles can potentially explain some of the phenotypic disparities seen between historically geographically distinct populations.
 The first report to link gene flow from Neanderthals to metabolic disease was in the Mexican and other Latin American populations in the form of SLC16A11 sequence variant (4). Mexican populations have nearly twice the prevalence of type 2 diabetes mellitus (5), and this report, a GWAS study, identified variants in the SLC16A11 and SLC16A13 genes to be novel risk loci associated with T2DM. Other GWAS looked at individuals primarily of European descent, and did not identify these loci. 5 variants (1 silent mutation, 4 missense SNPs) at the A11 locus, showing strong linkage disequilibrium, were strongly associated with T2DM (even after adjustments for age/BMI), and co-segregated into a common haplotype that is common amongst individuals of Latin American ancestry. The variant has also been shown to alter lipid metabolism in HeLa cells, increasing intracellular triacylglycerol levels. What's most interesting about this finding is how rare the 5SNP haplotype is throughout populations of European and African ancestry, while being found in the genome of a Neanderthal from the Denisova Cave (but not found in the reference sequences for Neanderthals or Denisovans). This 73kb spanning haplotype is nearly identical for both Neanderthals and individuals in the 1000 Genomes Project that are homozygous for the 5 SNPs. The authors conclude that it is highly likely that the haplotype was introduced into modern humans via archaic admixture; thus, gene flow from Neandethals conferred a relatively higher risk of developing T2DM (2.1 years earlier, at a .9 lower BMI) to this population.
The first report to link gene flow from Neanderthals to metabolic disease was in the Mexican and other Latin American populations in the form of SLC16A11 sequence variant (4). Mexican populations have nearly twice the prevalence of type 2 diabetes mellitus (5), and this report, a GWAS study, identified variants in the SLC16A11 and SLC16A13 genes to be novel risk loci associated with T2DM. Other GWAS looked at individuals primarily of European descent, and did not identify these loci. 5 variants (1 silent mutation, 4 missense SNPs) at the A11 locus, showing strong linkage disequilibrium, were strongly associated with T2DM (even after adjustments for age/BMI), and co-segregated into a common haplotype that is common amongst individuals of Latin American ancestry. The variant has also been shown to alter lipid metabolism in HeLa cells, increasing intracellular triacylglycerol levels. What's most interesting about this finding is how rare the 5SNP haplotype is throughout populations of European and African ancestry, while being found in the genome of a Neanderthal from the Denisova Cave (but not found in the reference sequences for Neanderthals or Denisovans). This 73kb spanning haplotype is nearly identical for both Neanderthals and individuals in the 1000 Genomes Project that are homozygous for the 5 SNPs. The authors conclude that it is highly likely that the haplotype was introduced into modern humans via archaic admixture; thus, gene flow from Neandethals conferred a relatively higher risk of developing T2DM (2.1 years earlier, at a .9 lower BMI) to this population. |
| Nature.com. GW=Genome Wide, LCP= Lipid Catabolism |
As much I think it'd be really interesting to conclude this is the result of gene flow, other options must be considered. It's clear that there's some selection on LCPs that are affecting gene expression + brain lipids concentrations in Europeans and Neanderthals, but we can't entirely assume this is from gene flow. Is this some sort of incomplete lineage sorting? Were these variants present in the most recent common ancestor of Neanderthals and humans, and driven to a higher frequency due to some (elusive) selective factor, living in the same environment? Until the 1000 genomes project gets a larger sample size, we can't entirely conclude that these variants aren't present in other populations at varying prevalences, drifting about unselected for. I'll be interested to see more paleoanthropologists weighing in on the potential environmental conditions that may have led to the positive selection on this haplotype. Having only looked at one body system (brain), I'd take every interpretation put forth with several grains of salt.
The authors controlled for environmental effects on metabolites by normalizing the metabolic divergence in the LCP term to the divergence based on all other metabolic pathways within the same population. However, looking at the metabolite categories that are effected most and match to divergent gene expression (trigylcerides, cholesterol esters, lecithin, ceramide, sphingomyelin), diet/nutrition likely play a major role in affecting these metabolites concentrations. One wonders how much of the noted divergence is due by nutrition (though I'm doubtful the trends seen would actually be significantly affected). Time to start hypothesizing how available nutrients in the shared European environment may have interacted with this haplotype! I personally find the choline stuff really interesting.
These analyses are prime examples of the need for focusing on identifying nutrient/metabolism-related haplotypes. A lot of individual gene variants grab the spotlight in nutrigenetics (ApoE4, Cytochrome P450 CYP, FADS, MTHFR), but it's more than likely that profound phenotypes have polygenic origins. As the study of nutrigenomics continues, i hope to see more evolutionary anthropology and population genetics playing a role.
I'm also excited to see what other variants may have flowed from ancestral hominids like Neanderthals, Denisovans, and these unknown hominids we're finding evidence of (7,8)... We need more studies of this Paleo Diet!
1. http://www.smithsonianmag.com/science-nature/homo-antecessor-common-ancestor-of-humans-and-neanderthals-143357767/?no-ist
2. http://www.pnas.org/content/early/2013/10/15/1302653110
3. http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1002947
4. http://www.nature.com/nature/journal/vaop/ncurrent/full/nature12828.html
5.http://www.ncbi.nlm.nih.gov/pubmed/20585724?dopt=Abstract&holding=npg
6. http://www.nature.com/ncomms/2014/140401/ncomms4584/full/ncomms4584.html
7. http://www.nature.com/nature/journal/v464/n7290/full/nature08976.html
8. http://www.upi.com/Science_News/2013/11/19/DNA-of-early-hominid-found-to-include-mystery-early-genes/UPI-70651384895737/
Comments
Post a Comment